MATERIAL & METHODS FOR TUMOR AND ATROPHY
The information presented in this methodological analysis was taken the Brain Web dataset . In this research methodological analysis, Brain Web dataset was utilised with different BMRI images. Based on standard tissue cleavage mask, Brain Web datasets had given the MRI encephalon images with undependable image quality and the datasets were excessively based on an anatomical construction of a normal encephalon, which had resulted from the undertakings of registering and preprocessing of 27 scans from the same single with cleavage. Different sorts of tissues were good identified in this dataset, both the types of tissue ranks “fuzzy” and “crisp” were assigned to each voxel. The sample encephalon MRI images from the Brain Web informations set were specified in the figure 4.1 given below.
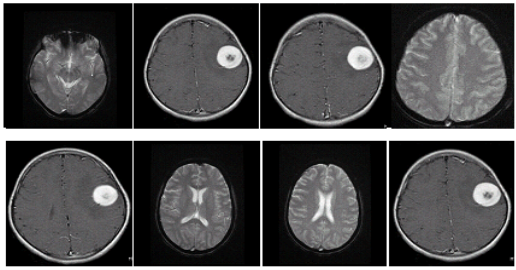
Fig 4.1 Sample images from Brain Web informations set
In this survey, the stuff was depended on the Brain Web informations. Brain images were taken for this survey ; it was examined and evaluated to observe the Brain tumour and Atrophy disease in the encephalon utilizing MRI. The methods adopted in the research were Artificial Intelligence and Neuro Fuzzy Classifier for BMRI image cleavage for encephalon tumour sensing and wasting diseases ; it was analyzed in this chapter 4.
The encephalon images utilized in the information set, which was used to measure the assorted images and its wasting degree and detected the highest wasting degree the images in the Brain Web dataset. Thus the public presentation analyzes of the encephalon images was used to gauge the following parametric quantities such as False Positive Rate, False Negative Rate, Sensitivity, Specificity and Accuracy, the overall public presentation of the system was determined. The basic count values such as True Positive ( TP ) , True Negative ( TN ) , False Positive ( FP ) and False Negative ( FN ) were used in this public presentation measures. Hence, the Brain Web images was included for the both the normal and unnatural images.
ARTIFICIAL INTELLIGENCE BASED BMRI IMAGE SEGMENTATION
Artificial Intelligence had been developed for scope of applications such as map estimate, optimisation, extraction, pattern sensing and classification. Particularly, they had been developed for encephalon images ; Features extraction, images cleavage, form and classification. Among the above applications, encephalon image cleavage was really indispensable as it plays a critical function for the high criterion of processing for encephalon tumour sensing in the medical images. Artificial Intelligence was besides used for assorted methods such as Back Propagation Networks, Radial Basis Function, Multi-Layer Perception, Self Organized Map ( SOM ) and Adaptive Resonance Theory. These nervous webs had been utilized for encephalon image cleavage. These techniques had been classified into provender frontward ( associatory ) and feedback ( car associative ) unreal nervous webs.
Artificial nervous webs had been demonstrated themselves as a proficient categorizer and were particularly good fitted for encephalon tumour classification. The back extension web was the of import tool in this survey. The input image was given to the web, which was non executable because the weight matrix was big. Another method was characteristics extraction from the given input images, which was used to develop the web. The back extension web was a supervised learning criterion for a provender frontward, multi layer nervous web that utilised sunburn sigmoid with predefined measure size to carry through preparation or supervised larning methods by utilizing the mistake rectification procedure and weight updating in this survey.
BACK PROPAGATION NETWORKS
Backward extension web was originated from the supervised acquisition method. It was a nonlinear abstraction of the squared mistake gradient descent supervised learning attack for updating the degree and weights of the nerve cells in a single-layer perceptual experience, extrapolated to feed frontward webs. Back Propagation needed the activation map, which was utilized by the unreal intelligence nodes or nerve cells, which was distinguishable from its ain derived function to a simple map.
The Back Propagation Networks had permitted to compare the gradient of the comparative mistake map to the concealed division and hence to cipher the weights of the concealed division for finding its value by gradient descent in the same manner as it to be estimated for the result subdivisions. Back propagated mistakes were theoretically obtained by using the classical concretion concatenation rule. In online supervision larning attack had estimated the weights of the unreal nervous web were upgraded at each and every input information point presentation. It had expended the resilient back extension version, which had employed merely the derivative map to put to death the weight upgrading.
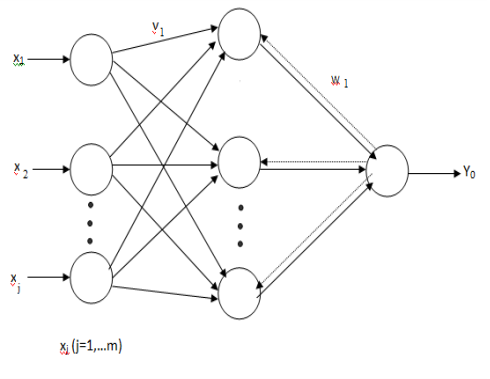
Fig 4.2 Back Propagation Network
The unreal nervous web was aimed for developing neither distinct value nor uninterrupted valued characteristics to measure the public presentation of the nervous web for altering inputs to acknowledge the encephalon tumour sensing. This methodological analysis had required low-level formatting of weights, mistakes of back extension and updating the weights. There was form or pre-structured acquisition and batch or pre-epoch supervised acquisition. In pattern larning methodological analysis, provided input images was patternized and so it was moved to propagated frontward, the mistake was estimated and back propagated and weights were besides updated. In batch acquisition methodological analysis, the whole set of developing webs weights were updated at the terminal and hence it had been demonstrated to the nervous webs. Thus the update of the weights was calculated merely after every era.
The nerve cell results were equated in the end product bed with the expected or direct results . The extension mistake had been calculated and so it was propagated through the nervous web and this determined value was utilized for the weight updating and rectification. The overall mistake was reduced at each and every province by utilizing the nerve cell weight accommodation. By and large the supervised acquisition rate had fixed what measure of the mensural mistake gradients had been utilized for the weight rectification. The accurate value of supervised larning rate was based on the mistake values. Some indication had been provided by mistake computation and noticed the old weight corrections. To better the truth in this methodological analysis, accelerate the nerve cells preparation for the nervous webs.
Artificial Neural Network developing attack was estimated by a group of conflicting necessity such as flexibleness, efficiency and simpleness . Simplicity was determined as the sum of the attempt needed to use the computational complexness. The form of Back Propagation Network ( BPN ) was displayed in figure 4.2. Flexibility was associated to the extendibility of the attack to develop assorted methodological analysis architectures and efficiency which referred the computational necessity for preparation and the success of the preparation phase. Thus the weight matrix job solution was viewed as a by-product of the preparation stages it was executed efficaciously.
EXPERIMENTAL RESULTS
In this proposed methodological analysis, all the encephalon MRI images were utilized in the experiment were in DICOM format and were developed from the original encephalon images. The size of the encephalon images utilized as 512 512 and were gathered from assorted patients. Hence, from these affected patients, 33 encephalon images were studied for this survey. The 20 dataset was trained and examined.
512 and were gathered from assorted patients. Hence, from these affected patients, 33 encephalon images were studied for this survey. The 20 dataset was trained and examined.
In pattern sensing web, which was referred as feed-backward web with tan-sigmoid transportation maps in both the concealed units and the end product units, was utilized. The nervous web had merely one end product neuron, because it had 20 input parametric quantities. The concealed units of nerve cells were 100 and the supervised acquisition degree was 0.1. The impulse constituent was 0.7 and overall eras were 400. The mistake was reduced by 0.0012 and the public presentation of the categorizer was analyzed by calculating truth of the images.
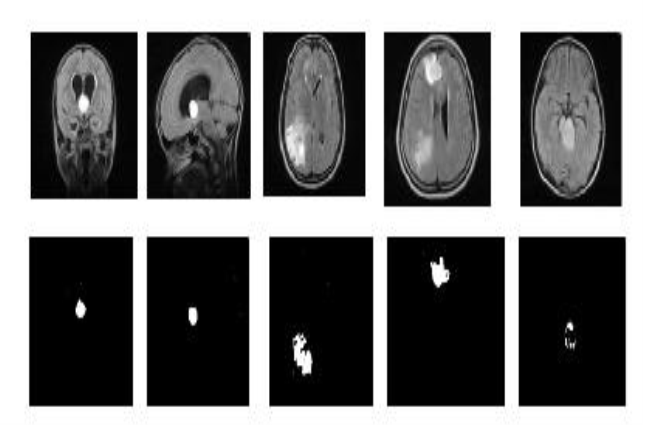
Fig 4.3 Detection of encephalon tumours
The form and strength characteristics of 33 encephalon images with tumours were calculated. The encephalon images were foremost classified into sub images and the stand foring characteristics were received utilizing Gabor method. The experimental results were displayed in the figure 4.3. The generalised characteristics of the encephalon tumours country for each MRI piece were fixed and were fed to Back Propagation Networks. Using the trained characteristics, encephalon tumour cleavage result for the same selected combinations of the extracted characteristics in the individual manner of piece was found. As the results, the encephalon tumour part was identified within a pixel size and preciseness degree was besides increased utilizing trained unreal impersonal webs.
In this methodological analysis, encephalon tumour sensing was using preprocessing measure and so Gabor characteristic extraction and eventually Back Propagation Networks classification was introduced. The stimulation consequence had presented the classification truth of 88 % . The system had been examined with assorted encephalon images. It was really of import to use for immense figure of patients information, which would besides farther heighten the preciseness degree of the system.
PHASES OF NFBS ( NEURO FUZZY BASED BMRI IMAGE SEGMENTATION )
Brain tissue cleavage from the MRI images was significance survey in the medical research field. The accurate cleavage of the normal every bit good as the unnatural tissues was the complex assignment in this procedure. Because of the incompatibility and trouble of unnatural tissues, MRI Brain Image Segmentation turned into more difficult process.
In this proposed methodological analysis, a technique was proposed for sectioning the abnormalcies such as Tumor and Atrophy in the MRI Brain images. By utilizing three phases such as characteristic extraction, categorization and cleavage were presented in this proposed methodological analysis. At first, the characteristics extraction was estimated by the undermentioned factors such as energy, information, homogeneousness, contrast and correlativity from MRI encephalon images were extracted. Subsequently, by using Neuro-Fuzzy classifier, the categorization procedure was carried out and for this phases, the characteristic set was specified as the input. From the result of categorization, the encephalon images were categorized into normal every bit good as abnormal. The farther process cleavage was performed harmonizing to this result merely.
The unnatural MRI images were segmented into unnatural tissues like Tumor and Atrophy utilizing Region Turning method. Using MATLAB platform the execution of the proposed technique was made. The experimentation was carried out on the MRI encephalon images by BrainWeb informations sets. The public presentation of proposed technique was assessed with the aid of the prosodies viz. FPR, FNR, specificity, sensitiveness and truth. Therefore, utilizing proposed technique with enhanced categorization, the unnatural tissues of MRI Brain images were segmented accurately.
Initially, the BMRI images were given as input to the proposed work and the characteristic sets were extracted from these input images. From these feature sets, the images were classified into two sorts of tissues – normal and unnatural utilizing the Neuro-Fuzzy classifier. Then the unnatural tissues Tumor and Atrophy were segmented utilizing Region Turning Method. The proposed work was illustrated in Fig. 4.4. Thus the phases of the proposed methodological analysis were demonstrated at a lower place such as given below:
- Feature set Extraction
- Neuro-Fuzzy classifier based Categorization
- Classified tissue’s Cleavage
FEATURE SET EXTRACTION:
In encephalon tumour sensing in the encephalon MRI images ; characteristic extraction was a curious signifier of dimensionality simplification. When the encephalon input information was given to an algorithm, which was really immense to be processed and it was mistrusted to be notoriously excess and so given input parametric quantities information had been translated into a minimized presented group of characteristics. Feature Extraction was utile in observing encephalon tumour where, it was exactly occurred and helpers in placing farther phases. Translation of the input information into the group of characteristics was called characteristic extraction.
In order to sort the given Brain MRI images, the characteristics from these MRI images were ab initio extracted. In this proposed work, the statistical characteristics such as Energy, Entropy, Homogeneity, Contrast and Correlation are extracted from these input BMRI images.
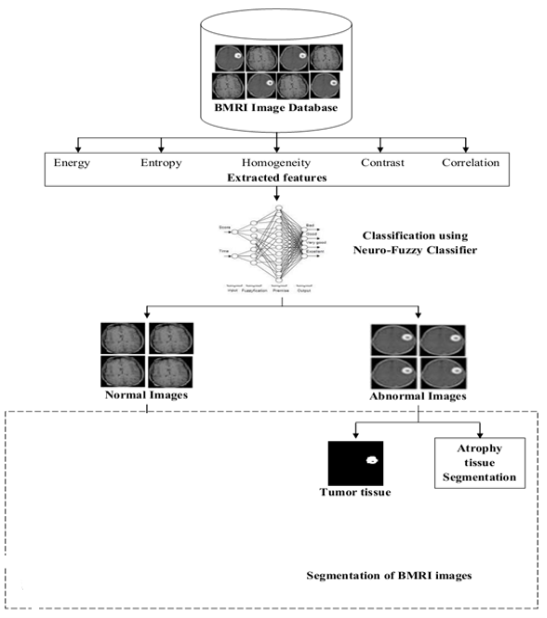
Fig 4.4 Proposed Neuro-Fuzzy based cleavage block diagram
CLASSIFICATION USING NEURO- FUZZY CLASSIFIER
The BMRI images were classified utilizing the Neuro-Fuzzy classifier. The extracted characteristics were given as the input to the Neuro-Fuzzy Classifier for sorting all the given BMRI images into 2 categories such as Normal BMRI images and Abnormal BMRI images. The Neuro-fuzzy system had a three-layered architectural design ; the undermentioned diagram fig.4.5 showed the basic construction of the neuro-fuzzy classifier system. Neuro-Fuzzy classifier was a fuzzy based system that was trained by a acquisition algorithm derived from Neural Networks. The acquisition algorithm merely performs on the local information and provided the local alterations in the fuzzy system. In general, a neuro-fuzzy system had generated really powerful solutions alternatively of utilizing the system components separately.
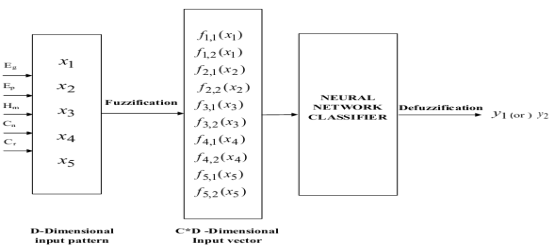
Fig 4.5 Architecture diagram for Neural Networks
Fig. 4.5 had displayed that proposed methodological analysis for cleavage of normal tissues utilizing improved machine larning attack. In this proposed work, it had used BrainWeb datasets for experimentation which included both normal and unnatural images. Energy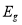 , Entropy
, Entropy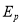 , Homogeneity
, Homogeneity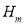 , Contrast
, Contrast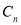 , and Correlation
, and Correlation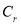 were the characteristics extracted from the encephalon MRI images.. These characteristics were so applied to a Neuro-fuzzy classifier that classifies the images into normal and unnatural. Proposed improved machine larning attack was a Neuro-fuzzy system that had a three-layered architectural design. This classifier was a fuzzy based system that was trained by a acquisition algorithm derived from nervous webs. The larning algorithm performs merely on the local information and provided the local alterations in the fuzzy system. In general, Neuro-fuzzy system had generated really powerful solutions when compared to the usage of system constituents separately.
were the characteristics extracted from the encephalon MRI images.. These characteristics were so applied to a Neuro-fuzzy classifier that classifies the images into normal and unnatural. Proposed improved machine larning attack was a Neuro-fuzzy system that had a three-layered architectural design. This classifier was a fuzzy based system that was trained by a acquisition algorithm derived from nervous webs. The larning algorithm performs merely on the local information and provided the local alterations in the fuzzy system. In general, Neuro-fuzzy system had generated really powerful solutions when compared to the usage of system constituents separately.
SEGMENTATION OF CLASSIFIED TISSUES
In medical image processing, cleavage dramas important function in implementing in encephalon images. Cleavage had been utile in different applications such as designation of tumour in the encephalon, ciphering tumour size and its volume, designation of the coronary cells in the angiograms, classification of the blood cells, encephalon images and so eventually in extraction of bosom images etc.
In by and large, it had been utile for sorting the images into anatomical country in few diligence for illustration musculuss, blood cells, castanetss, and blood vass, mean while few had been in the pathological country for illustration multiple induration, malignant neoplastic disease cells, and tissues malformations. To separate images into country, which were similar to few features and it had basic aim in the cleavage methods.
The chief end of the cleavage was to detach the full images accurately into sub countries sing white affair, Cerebrospinal fluid and grey affair of the encephalon MRI. Particularly, while including the measure of the neurological diseases such as Alzheimer’s upset and Multiple induration, the size and its volumes modified harmonizing to the entire encephalon, grey affair and white affair was offered as chief part in axonal loss and neural.
The Brain MRI images were categorized harmonizing to the neuro-fuzzy classifier, subsequently the encephalon mages were made available in this two different images merely such as pathological images or unnatural and normal. In general normal images used to hold the normal tissues such as cerebrospinal fluid, grey affair and white affair, which were utilized for cleavage.
The chief measure in sectioning the encephalon images, at first the images as to be preprocessed and so it was performed merely in the normal encephalon images, it was applicable for unnatural encephalon images. Finally, it was elementary to settle the cerebrospinal fluid tissue of the normal encephalon images in the part that covered the cerebral mantle by the use of the pre-processing attack. To section the encephalon tissues, it had gone through the preprocess phase and so it was shifted to the categorization of the images efficaciously.





