Gene look profiles on foretelling protein interaction web and exploring of new interventions for lung malignant neoplastic disease
Purposvitamin E:In the present survey, we aimed to research disease-associated cistrons and their maps in lung malignant neoplastic disease.Methods:Gene look profile GSE4115 was downloaded from Gene Expression Omnibus database. Entire 97 lung malignant neoplastic disease and 90 next non-tumor lung tissue ( normal ) samples were used to place the differentially expressed cistrons ( DEGs ) by paired t-text and discrepancy analysis in spectral angle plotter ( SAM ) bundle in R. Gene Ontology ( GO ) functional enrichment analysis of DEGs were performed with DAVID, followed by building of protein-protein interaction ( PPI ) web from HPRD. Finally, web faculties were analyzed by the MCODE algorithm to observe protein composites in the PPI web.Consequences:Entire 3102 cistrons were identified as DEGs at FDR & A ; lt ; 0.05, including 1146 down-regulated and 1956 up-regulated DEGs. Go functional enrichment analysis revealed that up-regulated DEGs chiefly participated in cell rhythm and intracellular related maps, and down-regulated DEGs might act upon cell maps. There were 39240 braces of PPIs in human obtained from HPRD databases, 3102 DEGs were mapped to this PPI web, in which 2429 braces of PPIs and 1342 cistrons were identified. With MCODE algorithm, 48 faculties were selected, including 5 corresponding faculties and 3 faculties with differences in cistron showing profiles. In add-on, three DGEs, FXR2, ARFGAP1 and ELAVL1 were discovered as possible lung malignant neoplastic disease related cistrons.Decision:The find of featured cistrons which were likely related to lung malignant neoplastic disease, has a great significance on analyzing mechanism, separating normal and malignant neoplastic disease tissues, and researching new interventions for lung malignant neoplastic disease.
Cardinal words:lung glandular cancer ; differentially expressed cistrons ; protein-protein interaction web ;
Introduction
As a sort of common malignant tumour, lung malignant neoplastic disease is a taking cause of malignant neoplastic disease decease worldwide [ 1 ] . The rates of lung malignant neoplastic disease happening in China and some Asiatic and African states are increasing [ 2-4 ] . Surgery has been used to handle lung malignant neoplastic disease historically and has been received positive experience in patients at the early phase [ 5-7 ] , and in phase III disease of lung malignant neoplastic disease [ 7-9 ] . Primary lung tumours were limited to bronchi pneumonic lobules, hence, early before lymphoglandula metastasis and distant metastasis, 5 old ages of endurance rate after the operation can be amounted to over 50 % . However, the high mortality rate is partially due to missing of effectual diagnostic method for the disease at the early phase [ 10 ] . As there is even no important symptom in early lung malignant neoplastic disease, diagnosing based on symptoms to observe lung malignant neoplastic disease is non really effectual. The first diagnosing can non be made until disease has already reached the metastatic stage ( phase a…? or a…? ) [ 11 ] . Consequently, early accurate diagnosing and therapy remain the of import missions of medical specialty research workers in the long periods from now on [ 12 ] .
These yearss, with the extremely development of bio-technology and invariably betterment of high-throughput showing ( HTS ) engineering, people return to the scientific survey of the nature of lung malignant neoplastic disease and its causes, procedures, development, and effects on the genome degree [ 13 ] . Gene look profiling utilizing microarrays is a robust and straightforward manner to analyze the molecular characteristics of different types and subtypes of malignant neoplastic disease at a system degree [ 14 ] . Recent researches base on the information of cistron look profiles to choose biomarkers associated with lung malignant neoplastic disease, uniting cistron look profiles with bio-networks ( PPI web ; signal web ; regulative web ; metabolic web ) , the complex pathogenesis of lung malignant neoplastic disease are analyzed [ 15, 16 ] . This simple technique has been proved to be helpful in accurate diagnosing of lung malignant neoplastic disease at the early phase [ 17 ] . It has been confirmed that the development of lung malignant neoplastic disease is a multi-step and multi-stage procedure in which many cistrons participate [ 11 ] . Thus, placing the differentially expressed cistrons ( DEGs ) between normal non-tumor and lung malignant neoplastic disease tissues can escalate the comprehension for the molecular mechanisms of lung malignant neoplastic disease happening and development [ 14 ] .
In this survey, we downloaded GSE4115, identified DEGs and performed functional enrichment analysis to derive the possible molecular mechanisms of lung malignant neoplastic disease. In add-on, we constructed protein-protein interaction ( PPI ) web and faculty analysis. By observing web faculties, the pathogenesis of lung malignant neoplastic disease was farther analyzed from modularity position.
Materials and methods
Datas preprocessing
The microarray informations of GSE4115 [ 18 ] were downloaded from Gene Expression Omnibus ( GEO ) database, the largest comprehensive public functional genomics informations depository. A sum of 187 french friess derived from bronchial epithelial tissue of tobacco users were available, including 90 normal samples ( without lung malignant neoplastic disease ) and 97 lung malignant neoplastic disease specimens ( with lung malignant neoplastic disease ) [ 19 ] . The probe-level informations were converted into the corresponding familial symbols, establishing on the relationship of the cistrons and the fiting investigations on platform GPL96. By taking the mean look value, the look values of all investigations for each cistron were convert to a individual value. .
Analysis of DEGs
After the cistron look was analysing by AffymetrixGenechip System ( Affymetrix, Santa Clara, CA ) , the SAM algorithm was applied straight. With t-text and discrepancy analysis, DEGs were identified. By commanding false find rate ( FDR ) with SAMr [ 20 ] bundle in R, SAM algorithm was used to avoid the multi-test job to cut down false positive consequences. FDR & A ; lt ; 0.05 was considered to be important DEGs.
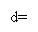
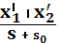 Relative difference ( vitamin D ) was computed as follow expression:
Relative difference ( vitamin D ) was computed as follow expression:
The d-statistics assay the comparative differences between cistron looks. Ten1’ represents the mean cistron look degree on status 1, while Ten2’ represents the 1 on status 2 and s is the discrepancy of the cistrons.
Functional enrichment analysis
The Gene Ontology ( GO ) functional and Kyoto Encyclopedia of Genes and Genomes ( KEGG ) pathways analysis were performed utilizing Database for Annotation Visualization and Integrated Discovery ( DAVID ) [ 21, 22 ] . With fisher accurate trial, the enrichment of a group of cistrons at each GO map node or KEGG tract was detected in the follow expression.
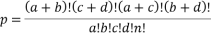
“a” represents DEGs in KEGG tract or GO cistron bunch ; “b” represents non DEGs but in KEGG tract or GO cistron bunch ; “c” represents DEGs but non in KEGG tract or GO cistron bunch ; “d” represents non DEGs and non in KEGG tract or GO cistron bunch.
PPI web building
As a protein database, HPRD ( Human Protein Reference Database ) [ 23 ] is accessible through the cyberspace. Comparing with other PPI databases like BIND [ 24 ] aˆ?DIP [ 25 ] aˆ?intAct [ 26 ] aˆ?STRING [ 27 ] and so on, informations about human PPI relationships and cistrons related to some diseases in HPRD were more abundant ( Fig.1 ; Fig2 ) . Therefore, the PPI from HPRD were collected for the building of differential protein interaction web. The DEGs were mapped to the HPRD database and screened important interactions. By incorporating these relationships, interaction web was constructed.
Designation of campaigner web molecules
The MCODE algorithm [ 28 ] was applied to observe protein composites in big PPI webs and was calculated utilizing a vertex-weighting strategy. The constellating coefficient CIwas used to mensurate ‘cliquishness ‘ of the vicinity of a vertex [ 29 ] via the expression: Degree centigradeI= 2n/ki ( ki-1 ) where qi denotes the figure of nodes which straight connected with nodes i, n denotes the figure of borders among these qis nodes. The maximal weight of a node was regarded as seed. After adding the node J which was over a certain threshold, the weight ratio ( Wj / W seed ) was searched out. A faculty was constructed by reiterating hunt until J does non run into the given threshold via the MCODE algorithm of plug-in clusterViz in CytoScape [ 30 ] . In this manner, the densest parts of the web are identified.
Consequences
SecondcreeningDEGs
Entire 3102 DEGs were selected at FDR & A ; lt ; 0.05 among normal and lung malignant neoplastic disease samples, including 1146 down-regulated and 1956 up-regulated DEGs.
Functional enrichment analysis
With FDR order, top 15 classs were selected. The functional enrichment of up- and down-regulated DEGs was shown in Table 1 and Table 2, severally. The consequences suggested that the up-regulated DEGs chiefly participated in cell rhythm and intracellular related maps, and the down-regulated cistrons might act upon cell maps like cellular adhesion and motility.
PPIweb building
Entire 39240 braces of PPIs in human were obtained from HPRD databases, titling the proteins with the names of the cryptography cistrons at the same clip. To find the map of them in lung malignant neoplastic disease, 3102 DEGs selected out of cistron showing profiles were mapped to this PPI web, placing the interactions of them to organize a sub-network named differential protein interaction web. There were 2429 braces of PPIs and 1342 cistrons in it ( Fig.3 ) .
Network faculty sensing and analysis
Entire 48 faculties were selected from PPI webs, of which there were 14 faculties contained more than 5 cistrons. To place faculties related to lung malignant neoplastic disease, 219 cistrons were downloaded from databases CancerResource. Mapped these cistrons to the faculties with more than 5 cistrons, 5 corresponding faculties ( Fig.4 ) and 3 faculties with differences in cistron showing profiles ( Fig.5 ) were identified. Disagreement likely indicated the cistrons might take to lung malignant neoplastic disease ; while understanding suggested the cistrons might influent the development of lung malignant neoplastic disease. Module 1 and 2 in Fig.4 suggested that merely cistrons related to lung malignant neoplastic disease were down-regulated, and cistrons related to lung malignant neoplastic disease which were down-regulated besides appeared in Fig.5. Faculties in it contained cistrons did non map to lung malignant neoplastic disease. In these three faculties, FXR2 ( delicate X mental deceleration, autosomal homolog 2 ) , ARFGAP1 ( ADP-ribosylation factor GTPase-activating protein 1 ) and ELAVL1 ( ELAV like RNA adhering protein 1 ) showed differences in look.
Discussion
As there is no important symptom in early-stage lung malignant neoplastic disease and it is difficult to observe by conventional thorax skiagraphy, it is pressing to develop fresh methods of diagnosing and therapy. Molecular biological science visible radiations the farther survey of the pathogenesis underlying this disease. In this survey, we combined cistron showing profiles and selected out 3102 DEGs with FDR & A ; lt ; 0.05. After the functional enrichment analysis was performed, PPI web was constructed, followed by analysing the faculties of web. Identifying the interactions of webs, 2429 brace of PPIs and 1342 cistrons in it were identified.
As is known, all molecules in cell functioned differentially but hand in glove in modulating the activities of cells [ 32, 33 ] . By building interaction web and analysing GO functional enrichment, we were able to larn that how the DEGs influent cell activities. From the consequences of PPI web building of up- and down-regulated cistrons, it may propose that the up-regulated DEGs chiefly participated in cell rhythm and intracellular related maps, while the down-regulated DEGs might influent cell maps like cellular adhesion and motility. It has been reported that cell rhythm is besides closely associated with DNA reproduction. Cell rhythm and DNA reproduction are related to unnatural fixs. These abnormalcies can do deformity growing, mental deceleration, and even take to malignant neoplastic disease [ 34 ] . Consequently, the instability of intracellular and extracellular maps takes portion in the oncoming and procedure of lung malignant neoplastic disease.
As for protein degree, PPI networks aid supply the cooperation connexions of proteins. Besides, the DEGs were associated with the development of assorted diseases. However, the manner how these DEGs were related to each other could non be analyzed straight from showing profiles. Consequently, by manner of building PPI webs of DEGs, observing and analysing web faculties, the pathogenesis of lung malignant neoplastic disease was possible to detect from the fundamental law of different faculties [ 35 ] . The activity of cell is largely composed of a web by otherwise interacted faculties: groups of cistrons co-regulated to respond to different state of affairss [ 36 ] . The differences between cistron showing profiles in faculties likely account for the development of lung malignant neoplastic disease.
Besides, possible cistrons ARFGAP1, ELAVL1 and FXR2 which were related to lung malignant neoplastic disease were discovered while comparing faculties contained common profiles. As the over-expression and invasive actions, ARFGAP1 plays of import functions in tumour and malignance invasion [ 37 ] . In add-on, ARFGAP1 has over-expressed in seven malignant neoplastic disease cell lines [ 38 ] . ELAV1, named HuR every bit good, has been reported to be closely related to lung malignant neoplastic disease in several articles. HuR has been found to be expressed in NSCLC ( non-small-cell lung malignant neoplastic disease ) and have close relationship with lymphangiogenesis and angiogenesis [ 39 ] . FXR2 is a RNA binging protein which is similar to FXR1 and FMRP. FXR1 protein has found the ability to modulate the look of tumour mortification factor at the post-transcriptional degree [ 41 ] . Therefore, there are important grounds to reason that FXR2 is a member of possible lung cancer-related cistrons.
In drumhead, the find of featured cistrons which were perchance related to the development and advancement of lung malignant neoplastic disease, is non merely supported by old surveies, but besides more reified than the researches. In add-on, it has a great significance on analyzing mechanism, probi nanogram and separating normal next with cancerous tissues, every bit good as researching better methods for diagnosing and interventions for lung malignant neoplastic disease. However, farther experiments are still needed to corroborate our consequence.
Acknowledge
We wish to show our warm thanks to Fenghe ( Shanghai ) Information Technology Co. , Ltd.Their thoughts and aid gave a valuable added dimension to our research.
Declaration
All human surveies have been approved by China Ethics Committee and performed in conformity with the ethical criterions.
Conflict of involvement
Figures and tabular arraies
Fig.1.Comparison of PPI relationships and protein capacity among 8 PPI databases
Fig.2. Comparison of cistron Numberss and disease related cistron Numberss among 8 PPI databases
Fig.3 Differential Protein Interaction NetworkThe ruddy nodes stand for and the green nodes stand for n down-regulated cistrons. ( p-value & A ; lt ; 0.05, adjusted p-value & A ; lt ; 0.05 )
Fig.4 5 Modules Mapped to Genes of Lung Cancer. Round nodes represented known cistrons of lung malignant neoplastic disease ; ruddy and green represented up- and down-regulated cistrons, severally.
Fig.5Three faculties with differences in showing profiles without mapping
Red base for up-regulated cistrons, green base for down-regulated 1s.
1





